
The production of cheap, durable and adaptable plastics has exploded in the last few decades as they infiltrate every part of our lives; but this once desirable polymer has a dark side. Plastics may take hundreds of years to degrade and with production at astronomic levels (globally over 350 million tons each year), plastic pollution is one of the most pressing environmental concerns facing the world today.
A staggering 150 million tons of plastic finds its way to landfill or is released into the environment annually and over 8 million tons are transported by rivers into the world’s oceans. Most of this does not degrade but simply breaks down into microparticles. These well-documented microplastics are found in ocean water, within marine animals and even deep inside the gastroenteric systems of humans. Plastic pollution is one of the most critical environmental issues facing humankind today and researchers have been busy looking for answers to this perplexing plastic problem.
Depolymerization: Solving the polymer recycling conundrum
Plastics are made of polymers – long chains created by repeating monomer blocks. Most of the widely used plastics are either thermoplastic or thermosetting. Thermoplastics, such as acrylic, polyamide and polyethylene, become soft and moldable at high temperatures and harden when cooled. This property makes them relatively easy to recycle because they can be softened and remolded into new products, although quality decline limits benefit. Thermosetting plastics, like polyurethane, epoxy resin and melamine resin, harden when heated and are almost impossible to recycle. The challenges faced by recycling both thermoplastics and thermosets mean that all plastics will eventually be destined to contribute to environmental contamination.
To attain real recycling, with subsequent reuse in new products, waste plastics need to be returned to their original monomers through a process called depolymerization. This is a critical technical advance needed to enable a global circular materials economy. In biological systems, complete depolymerization to monomers can be necessary for microbial uptake and growth.
To achieve depolymerization, scientists have looked to nature, searching for microbial enzymes that can break down plastics. In 2012, researchers at Osaka University discovered an enzyme in a compost heap that can break down one of the world’s most used plastics: polyethylene terephthalate (PET, CAS Registry Number 25038-59-9, formula (C10H8O4)n).
The enzyme, known as leaf-branch compost cutinase (LLC), breaks the bonds between PET monomers, but it is intolerant to the 65°C softening temperature of PET, denaturing after a few days of working at this temperature and limiting its industrial practicability. Since depolymerization can only take place in molten plastic, enzymes must be stable at increased temperatures
Double-action PET depolymerization from a little-known soil bacterium
PET is a thermoplastic and one of the most widely used polyesters. Worldwide PET production grew from 42 million tons in 2014 to 50 million tons in 2016 and is set to reach 87 million tons by 2022.
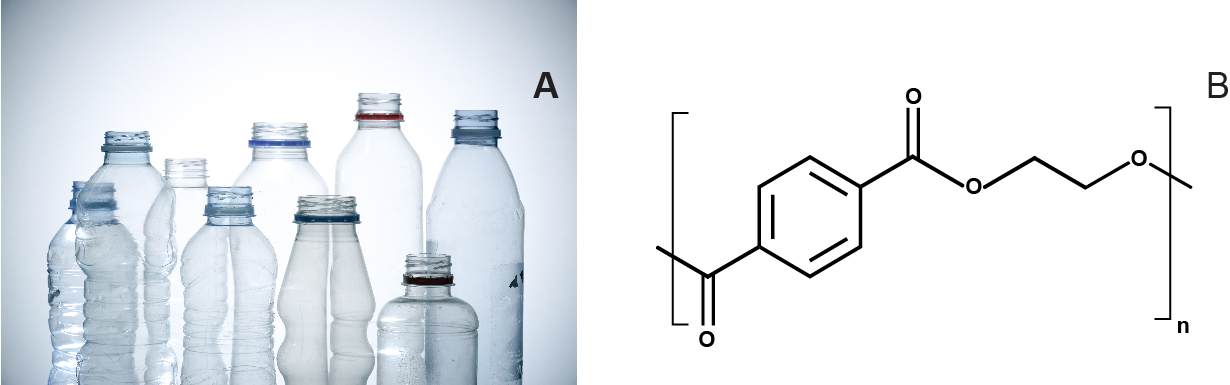
This synthetic polymer is manufactured from petroleum-derived terephthalic acid (TPA) and ethylene glycol (EG). PET is a versatile polymer that can be made transparent, opaque, or white in color, depending on its crystal structure and particle size (Fig. 1). It is widely used to produce clothing fibers and containers, including water bottles, and non-oriented PET can be thermoformed (or molded) to manufacture other packaging products, such as blister packs1. Finding an effective way to depolymerize PET will be an important milestone in the journey towards true plastic recycling and resulting environmental protection.
PET biodegradation has been extensively studied because esterase enzymes (enzymes that split esters into an acid and an alcohol) are abundant in nature2. Reports on the biological degradation of PET or its utilization to support microbial growth are, however, infrequent. Some organisms from the filamentous fungi group, Fusarium oxysporum and Fusarium solani, have been grown on a mineral medium containing PET yarns3.
In 2016, Yoshida et al4 reported the discovery and characterization of the soil bacterium strain, Ideonella sakaiensis 201-F6, found growing in PET-contaminated sediment near a plastic recycling facility in Japan. This gram-negative, aerobic, rod-shaped bacterium has the remarkable ability to use PET as its major carbon and energy source for growth.
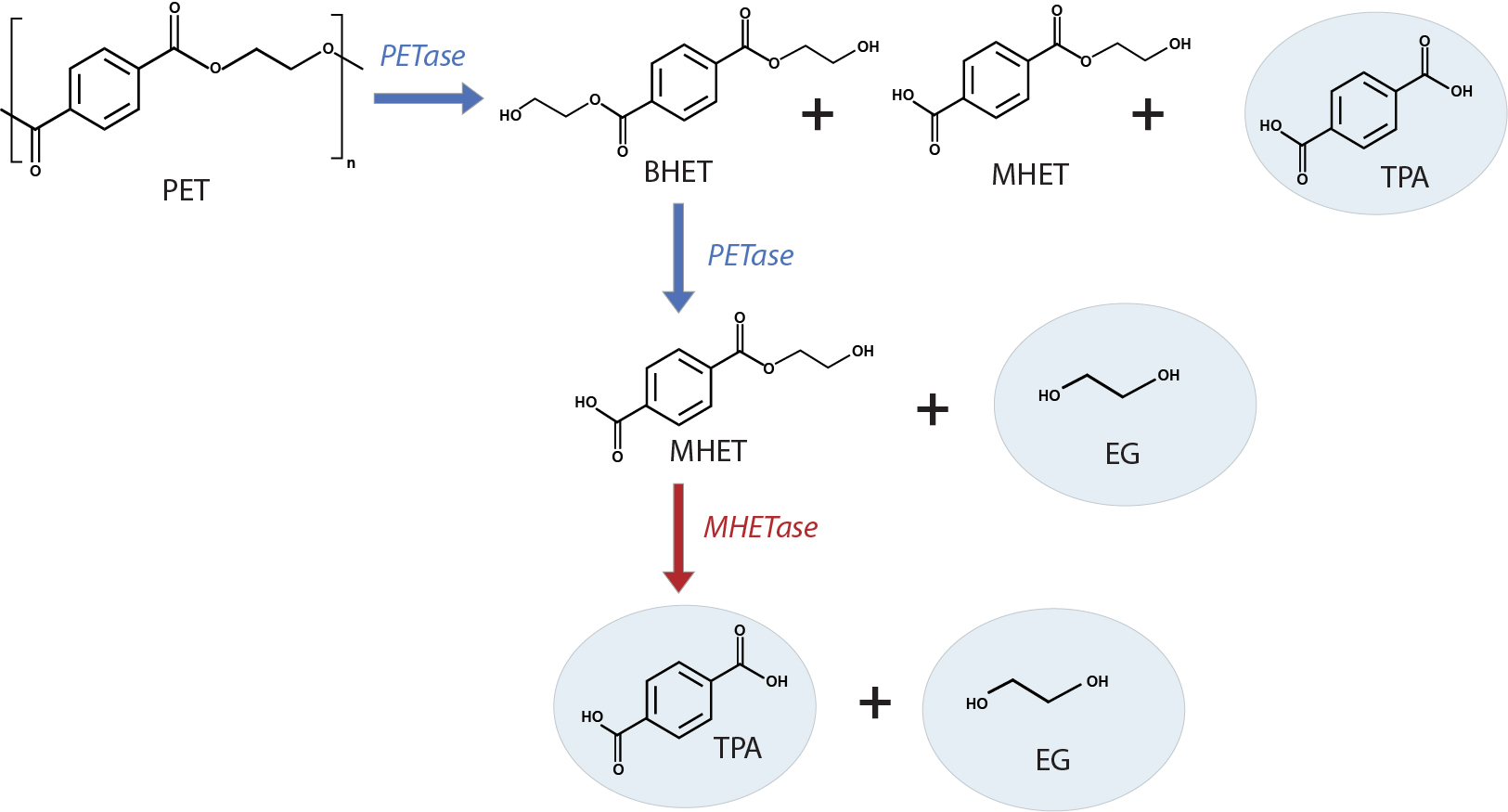
I.sakaiensis employs a two-enzyme system to depolymerize PET to its building blocks, TPA and EG, which are further catabolized to a carbon and energy source. One of the two enzymes, ISF6_4831 protein, hydrolyzes and breaks ester linkages. With a preference for aromatic rather than aliphatic esters, and a specific inclination towards PET, it is designated as a PET hydrolase (PETase). The PETase enzyme in I. sakaiensis is a cutinase-like serine hydrolase that attacks the PET polymer, releasing bis(2-hydroxyethyl) terephthalate (BHET), mono(2-hydroxyethyl) terephthalate (MHET) and TPA. PETase further cleaves BHET to MHET and EG. The second enzyme, ISF6_0224 protein, MHET hydrolase (MHETase), further hydrolyzes the soluble MHET to produce TPA and EG (Fig. 2). Both enzymes are required, likely synergistically, to enzymatically convert PET into its two environmentally benign monomers, TPA and EG4, making it possible to fully recycle PET.
PETase mutant’s supersize PET degradation capability
Sequence and structural studies of PETase enzyme highlight similarities to cutinase enzymes, evolved by many bacteria to break down cutin, a natural, waxy polymer that forms part of the protective cuticle in many plants. Crystal structural analysis and biochemical tests reveal that the PETase in I.sakaiensis 2 has an open active-site architecture in the binding site and that it likely operates along the canonical serine hydrolase catalytic mechanism5.
Based on structural modifications in PETase and a homologous cutinase active-site cleft, PETase variants have now been produced and tested for PET degradation, including a mutant with a double mutation distal to the catalytic center. This area is hypothesized to be capable of amending important substrate-binding interactions6. This double mutant, based on cutinase architecture, was found to exhibit enhanced PET degradation capacity relative to wild-type PETase6 and a patent has now been filed7.
By narrowing the binding cleft via the mutation of two active-site residues in cutinases, researchers observed improved PET degradation, suggesting that PETase does not exhibit the optimum structure for degradation of crystalline PET, despite evolving in a PET-rich environment. The mutant enzyme takes just a few days to start breaking down the plastic – significantly faster than the centuries it takes in the oceans.
From double mutant to a double enzyme cocktail
When MHETase enzyme is added to the reaction, the enzyme mixture breaks down PET twice as fast as PETase on its own. The degradation trend observed within the tested enzyme loadings range shows increasing levels of constituent monomers as the concentration of both enzymes increases. This indicates that the reactions are enzyme, rather than substrate, limited. The synergy analysis also indicates that degradation rates increase with PETase loading and that the presence of MHETase, even at low concentrations relative to PETase, improves total degradation. The current experiments do not indicate an optimal ratio of PETase to MHETase.
Creating a super-enzyme triples PET degradation
In further experiments exploring the properties and scope of PET degradation, researchers have engineered a new super enzyme by stitching MHETase and PETase into one long chain. The chimeric proteins covalently linking the C terminus of MHETase to the N terminus of PETase, using flexible glycine-serine linkers, were generated and assayed for degradation of amorphous PET (Fig. 3). When comparing the extent of degradation achieved by different enzymes, the chimeric proteins outperform both PETase and MHETase, as well as the unlinked enzyme mixture.
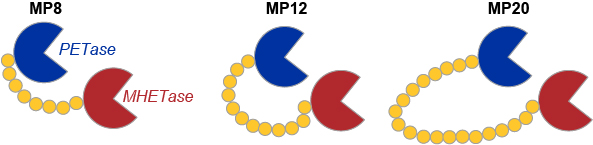
Interestingly, the super-enzyme not only depolymerizes PET but also works on PEF (polyethylene furanoate), a sugar-based bioplastic used in beer bottles.
The enzymatic deconstruction of some natural polymers, such as cellulose and chitin, is achieved in nature by mixtures of synergistically acting enzymes secreted from microbes8. These natural microbial systems have evolved over time to optimally degrade such polymers. It appears that some soil bacteria, such as I. sakaiensis, have evolved in a similar way to utilize a polyester substrate with a two-enzyme system4,9. Unlike natural degradation, which can take centuries, the super-enzyme can convert PET back to its monomers in just a few days, although this is still too slow to be commercially viable.
Endless recycling with plastic degradation
By converting PET back to its original, monomer building blocks, the super-enzyme would allow plastics to be made and reused endlessly, reducing dependence on fossil resources. And the breakthroughs don’t stop here…
In 2020, a major advance saw scientists identifying another enzyme that could degrade PET in just 10 hours10. The research screened a large variety of bacteria and enzymes for potential candidates, including the leaf-branch compost cutinase, LCC, that was first discovered in 2012. Hundreds of mutant PET hydrolase enzymes were then produced by varying amino acids at the binding site and improving thermal stability. Bacterial mutants were then screened to identify efficient PET decomposers. After running this process for multiple rounds, a mutant enzyme was isolated that is 10,000 times more efficient in degrading PET than the native LCC. It is also stable at 72°C, close to the melting temperature of PET. This finding contributes significantly towards attaining the infinite recycling of PET and is already at a pilot industrial stage10.
We are only seeing the tip of the iceberg of possibilities that these microorganisms and their enzymes can offer. Most plastics are derived from fossil fuels, finite in their creation yet ubiquitous in our environment. Plastic pollution will continue to be a growing problem unless we can find a way to form a circular economy. In just a few decades, we will be unable to produce the plastic items on which we depend unless we can find a way to recycle the waste that already exists. Traditional recycling is not effective or sustainable and, unless we can reduce plastics to their monomer constituents on an industrial scale, we cannot hope to solve this problem. Thankfully, with the help of nature, some skillful evolution and a dash of scientific ingenuity, there is a hope that this problem might be solved.
Interested in learning more about ‘green’ innovations?
In the hunt for novel species to drive innovation, nowhere contains greater biodiversity than the rainforests of Brazil. Learn how data is being collected and mined for public use and how this might help to protect the 15-20% of the Earth’s biodiversity.
(Read Organizing Vital Data to Unlock Innovation from Brazil’s Biodiversity)
REFERENCES
(1) Pasbrig, E.; Claessens, P.; Walker, R. I.; Walker, R. Peelable cover film for pharmaceutical packaging, e.g. blister packs, comprises paper, aluminum foil or heat-resistant plastic, a layer of special plastic film, mesh or fabric, a layer of aluminum foil and a heat-sealing layer. EP1767347-A1; WO2007038488-A2; EP1928654-A2; AU2006294788-A1; US2008251411-A1; CN101316702-A; CA2623586-A1; JP2009509874-W; TW200727887-A; MX2008004201-A1; IN200801248-P2; ZA200802826-A; BR200616412-A2; WO2007038488-A3; EP1928654-A4.
(2) Han, X.; Liu, W. D.; Huang, J. W.; Ma, J. T.; Zheng, Y. Y.; Ko, T. P.; Xu, L. M.; Cheng, Y. S.; Chen, C. C.; Guo, R. T., Structural insight into catalytic mechanism of PET hydrolase. Nature Communications 2017, 8. DOI: 10.1038/s41467-017-02255-z
(3) Nimchua, T.; Eveleigh, D. E.; Sangwatanaroj, U.; Punnapayak, H., Screening of tropical fungi producing polyethylene terephthalate-hydrolyzing enzyme for fabric modification. J. Ind. Microbiol. Biotechnol. 2008, 35 (8), 843-850. DOI: 10.1007/s10295-008-0356-3
(4) Yoshida, S.; Hiraga, K.; Takehana, T.; Taniguchi, I.; Yamaji, H.; Maeda, Y.; Toyohara, K.; Miyamoto, K.; Kimura, Y.; Oda, K., A bacterium that degrades and assimilates poly(ethylene terephthalate). Science 2016, 351 (6278), 1196-1199. DOI: 10.1126/science.aad6359
(5) Rauwerdink, A.; Kazlauskas, R. J., How the Same Core Catalytic Machinery Catalyzes 17 Different Reactions: the Serine-Histidine-Aspartate Catalytic Triad of alpha/beta-Hydrolase Fold Enzymes. Acs Catalysis 2015, 5 (10), 6153-6176. DOI: 10.1021/acscatal.5b01539
(6) Austin, H. P.; Allen, M. D.; Donohoe, B. S.; Rorrer, N. A.; Kearns, F. L.; Silveira, R. L.; Pollard, B. C.; Dominick, G.; Duman, R.; El Omari, K.; Mykhaylyk, V.; Wagner, A.; Michener, W. E.; Amore, A.; Skaf, M. S.; Crowley, M. F.; Thorne, A. W.; Johnson, C. W.; Woodcock, H. L.; McGeehan, J. E.; Beckham, G. T., Characterization and engineering of a plastic-degrading aromatic polyesterase. Proc. Natl. Acad. Sci. U. S. A. 2018, 115 (19), E4350-E4357. DOI: 10.1073/pnas.1718804115
(7) Beckham, G. T.; Johnson, C. W.; Donohoe, B. S.; Rorrer, N.; McGeehan, J. E.; Austin, H. P.; Allen, M. D. New modified polyethylene terephthalate -digesting enzyme comprising amino acid mutation of an active site residue, is used to degrade a polymer e.g. polyester, aromatic polymer or semi-aromatic polymer and polyethylene terephthalate. WO2019168811-A1.
(8) Payne, C. M.; Knott, B. C.; Mayes, H. B.; Hansson, H.; Himmel, M. E.; Sandgren, M.; Stahlberg, J.; Beckham, G. T., Fungal Cellulases. Chem. Rev. 2015, 115 (3), 1308-1448. DOI: 10.1021/cr500351c
(9) Taniguchi, I.; Yoshida, S.; Hiraga, K.; Miyamoto, K.; Kimura, Y.; Oda, K., Biodegradation of PET: Current Status and Application Aspects. Acs Catalysis 2019, 9 (5), 4089-4105. DOI: 10.1021/acscatal.8b05171
(10) Tournier, V.; Topham, C. M.; Gilles, A.; David, B.; Folgoas, C.; Moya-Leclair, E.; Kamionka, E.; Desrousseaux, M. L.; Texier, H.; Gavalda, S.; Cot, M.; Guémard, E.; Dalibey, M.; Nomme, J.; Cioci, G.; Barbe, S.; Chateau, M.; André, I.; Duquesne, S.; Marty, A., An engineered PET depolymerase to break down and recycle plastic bottles. Nature 2020, 580 (7802), 216-219. DOI: 10.1038/s41586-020-2149-4
Gain new perspectives for faster progress directly to your inbox.




